标题:NCBI LocusLink知识介绍和使用
浏览次数:1602
NCBI LocusLink知识介绍和使用
位点链接(LocusLink)提供一个单一查询界面来找到某一个遗传位点的序列和描述性信息。它展现官方命名,别名,序列登录,表型,EC号码,MIM号码,UniGene聚类,同源,图谱位点,和相关的网站信息。
序列等级包括一个GenBank为位点登录的子集,以及一种新的类型,NCBI参考序列(RefSeq)。RefSeq纪录是根据这样的步骤建立。参考FAQ页面获得更多的信息。
数据可以这样访问,通过单击页面顶端的字母来浏览由符号排序的位点,或者通过一个查询进入搜索表格。替代字母(*)的使用是被支持的。更多的信息和查询技巧可以在帮助文档里查到。
目前的范围是果蝇,人类,小鼠,大鼠,和斑马鱼。
LocusLink Frequently Asked Questions
我如何从一个位点记录到STS数据?
链接到STS记录并不被从LocusLink浏览列表上直接支持。但是,报告页面上的Map部分可能包含STS标记,是用电子PCR计算出来的,用来发现代表这个基因的cDNA。另外,如果一个UniGene链接(在浏览页面上,UniGene: Hs.00000在报告页面上)是被显示的,那么去UniGene用该聚类的序列确定STS。
我如何从一个位点记录到含有这个基因的图谱?
在人类和果蝇基因组的浏览列表上,Position栏提供了一个链接到一个恰当的图谱观察页面。在那里Display设置功能能够被用来控制被显示的图谱和区域。另外,如果一个UniGene链接(在浏览页面上,UniGene: Hs.00000在报告页面上)是被显示的,试着用UniGene来确定在放射杂交名单上有没有STS定位数据。或者一个OMIM链接(在浏览列表上,OMIM: 000000在报告页面上)被显示,那么去OMIM看看一个基因图谱记录是否存在。从报告页面,链接到Entrez图谱浏览器产生一个查询到细胞遗传学和序列基因图谱。对于大鼠和小鼠基因组,更多的图谱信息可能可以在合适的特定基因组数据库中找到。
一个UniGene链接是如何操作的?有时候,我不能看到一个链接,有时候我可以看到更多。
这是一个临时的情况。我们的目的是改善UniGene和LocusLink之间的对应,这样任何一个同序列数据锚定的基因,只有一个UniGene聚类对应。
如果我知道一个基因的登录号,而LocusLink却显示没有登录号,我如何更正这个遗漏?
我们鼓励你的提议。请发email给NCBI服务工作台,并提供你的名字和联系信息,以及你认为序列对应的登录号和位点ID。
我知道对应一个基因或蛋白有比我看到的要多的登录号。为什么?
LocusLink不是设计来报告一个给定基因或 蛋白所有发表的序列的登录号的。LocusLink的目的是提供一套已知的代表序列。其它工具如:Entrez“相关序列”或BLAST可以用来检索更多 的记录。LocusLink报告的登录号的数目和特定的例子取决于这个基因是否是一个临时的或已经被检验过的RefSeq记录,或者是否是RefSeq。
如果没有RefSeq记录,每一个登录号只有一个被建议代表这个基因,没有其它工具被用过来确定其它的相关序列。
如果RefSeq记录是在临时分类里,注解有完整编码区的附加的记录将被列出,编码区将不超过截断的核苷酸不匹配水平。
如果是检验过的,用来产生组装序列的那些序列被详细的报告。其它代表这个基因的GenBank记录也被列出,但是,这个列表并非是完全的。
我如和才能将一个登录号列表转化成位点ID的值?
一个将登录号转化成位点ID值的方法是使用文件 ftp://ncbi.nlm.nih.gov/refseq/LocusLink/loc2acc。这个每日更新的文件包含了引证在ftp: //ncbi.nlm.nih.gov/refseq/LocusLink/README.中的值。
LocusLink多久更新?
文件每周更新一次,过每个周末。统计页面将提供文件刷新的最近日期。
符号和基因名字是如何选择用于LocusLink和RefSeq记录的?
LocusLink和RefSeq都是是用对基因组合适的由命名委员会建立的基因符号和基因名字。在一个LocusLink报告中,符号和名字被在横幅下报告。
官方基因符号和命名。在一个RefSeq记录中,符号是以LOCUS和以特性gene=来报告的,名字是在DEFINITION中是用。
如果一个官方符号还没有被指定,那么一个暂时的符号和名字被人为的选择。
在形成一个检验过的RefSeq记录的分析过程中,可选的基因符号,基因名字,和蛋白名字被工作人员加入。
什么样的引文被选来用于LocusLink和RefSeq记录?
通过PubMed详细的链接到的引文由我们的工作人员选出,对于人类,这些引文主要对应人类基因命名委员会和OMIM。这些引文通常报告基因发现和定位。在形成一个检验过的RefSeq记录的分析过程中,有些引文记录了功能和一个基因的表达以及它的产物也被加入。
LocusLink报告和RefSeq记录并非准备报告所有的与这个基因相关的引文或它的蛋白产物。它的目的是提供一套引文用来提供一些背景信息。这些引文,以及那些被包含在OMIM报告中的,可以用PubMed的Related articles功能来发现相关的文章。
最近发表的文章:
• Introducing RefSeq and LocusLink: curated human genome resources at the NCBI. Pruitt KD, Katz KS, Sicotte H, Maglott DR Trends Genet. 2000 Jan;16(1):44-47.
• NCBI's LocusLink and RefSeq Maglott DR, Katz KS, Sicotte H, Pruitt KD Nucleic Acids Res 2000 Jan 1;28(1):126-128
How to Use LocusLink
1) What is LocusLink?
2) How does LocusLink relate to PubMed, RefSeq, and other NCBI databases?
3) How do I use LocusLink?
4) What do the search results mean?
5) Quick Quiz
1) What is LocusLink?
LocusLink (http://www.ncbi.nlm.nih.gov/LocusLink) is a National Center for Biotechnology Information (NCBI) online resource. It is principally intended for use by graduate students and professional researchers in the biomedical sciences. It is designed to bring together related information on genetic loci and gene products from several sources. LocusLink provides a central point of access for basic biomedical information and molecular data for genes, transcripts, and proteins from model organisms, currently including human, rat, mouse, fruit fly, and zebrafish.
2) How does LocusLink relate to PubMed, RefSeq, and other NCBI databases?
NCBI has a large and growing number of search tools for biologists to obtain information. A few of these include:
PubMed: a searchable biomedical literature citation index. For a given genetic locus, LocusLink leads directly to a short list of PubMed citations for that gene. (This list usually includes reports pertaining to central genetic or molecular biological discoveries, and to reports on disease-causing alleles, for the gene in question.)
RefSeq: Another new NCBI database, RefSeq (Reference Sequence) entries are intended to serve as ""authority files"" for genetic sequence information. For a given genetic open reading frame, RefSeq provides a curated file on the gene sequence and its transcriptional and translational processing (where available). An professional review process helps to ensure the biological accuracy of these authority files. RefSeq files are accessible directly from the LocusLink entry for the genetic locus in question.
OMIM (Online Mendelian Inheritance in Man): a database of human genes and genetic diseases, including knowledge of their molecular and physiological roles and causes. The writeups for genetic loci and their roles in physiology are often extensive and are frequently updated. OMIM files are accessible directly from the LocusLink entry for the genetic locus in question.
GenBank, Protein Database, Homologene, UniGene, genetic variations database (single nucleotide polymorphisms): links to gene-specific information from each of these databases are directly available from the LocusLink entry for the genetic locus in question.
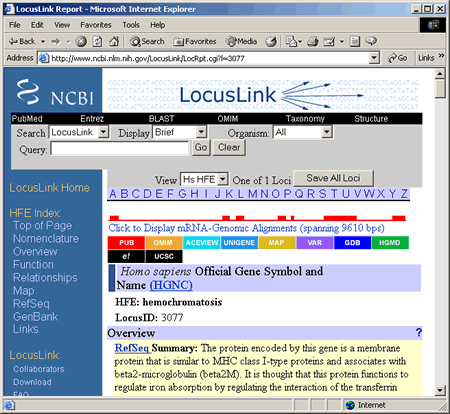
3) How do I use LocusLink?
Go to the LocusLink home page: http://www.ncbi.nlm.nih.gov/LocusLink. Although an alphabetical list of entries is available, LocusLink can be most easily searched using the query box at the top of the page. Users can enter a wide variety of terms, for example: gene name or gene symbol (e.g., SDHA), protein name (succinate dehydrogenase flavoprotein), protein symbol (SDH), EC (Enzyme Commission) number (1.3.5.1), and disease states (Leigh syndrome). Type in your search query into the ""Query:"" box, then press ""go"".
What data is LocusLink searching?
LocusLink searches the full text of the files within its database for the individual terms queried.
Can I do truncation searches?
Yes. You can use the asterisk as a truncation symbol (e.g., search term: succin*) to obtain, in this example, information on a variety of genetic loci involved in metabolism of succinate or succinyl-derivatives.
Can I do Boolean (AND, OR, NOT) searches?
If multiple terms are entered (e.g., succinate dehydrogenase) the search engine automatically searched for files containing both words (succinate and dehydrogenase) in the file. Searches can also be constructed using the terms AND, OR (to find files containing both or either search terms), and NOT (to find files containing the first but not the second term). Presently, however, proximity searches (succinate within “x” words of dehydrogenase), searches on phrases (""succinate dehydrogenase"" where the two words must be immediately adjacent), and synonyms are not fully supported.
4) What do the search results mean?
On the results page, first note that the number of entries returned is given. If you get no results, refer to the ""help"" section, linked in the left-hand bar on the page. If you do get hits, you can explore the NCBI site databases in a number of ways, depending on the data available for a given locus. These will be listed in a table with the headings ""LocusID"", ""Org"", ""Symbol"", ""Description"", ""Position"", and ""Links."" Each of these is described below:
Click on the blue ""LocusID"" number to get an overview description of the gene locus and its function,. (This is the unique database number for the entry.)
The ""Org"" column lists abbreviations for the organism that the gene was isolated from (Hs = Homo sapiens, Mm = Mus musculus, Dm = Drosophila melanogaster, etc.)
The ""Symbol"" column lists the common gene abbreviation for the locus.
""Description"" is a brief explanation of the function of the locus.
The ""Position"" column gives the chromosomal map location of the genetic locus. Clicking the blue entry links to a visual chromosomal map with the gene marked on it.
The rainbow-colored ""Links"" column gives links to several other NCBI databases:
“P” PubMed
“O” Online Mendelian Inheritance in Man (OMIM)
“R” RefSeq database
“G” GenBank database
“P” Protein database
“H” Homologene database
“U” Unigene database
“V” Variation data: single nucelotide polymorphism (SNP) database
If you can't find the genetic information you're looking for using LocusLink (or don’t understand why you found what you did!), please feel free to contact University of Illinois Library graduate assistant Kevin Messner (krmessne@uiuc.edu), or visit the reference desk of your biomedical library.
5) Quick Quiz:
a) How can I find a picture of the human chromosome map location of the SOD1 gene via LocusLink (http://www.ncbi.nlm.nih.gov/LocusLink)?
Answer: Search LocusLink for SOD1, then click on the ""Position"" link for the Homo sapiens (Hs) entry in the results table to bring up a chromosomal map with the gene marked.
b) Where can I find a basic description of function of the SOD1 gene and protein, and perhaps a description of genetic diseases it is related to?
Answer: Use LocusLink, then on the results page click on the OMIM button (orange button with an ""O"") for the SOD1 entry.
c) Using LocusLink (http://www.ncbi.nlm.nih.gov/LocusLink), can I find any known single nucleotide polymorphisms for the SOD1 gene? How about GenBank entries for the gene?
Answer: On the results page for SOD1, click on the V (""variations"") to reach the SNP database entries for the SOD1 gene. Click on the G to access GenBank records.
d) When I search for SOD1, I also get a hit on a ""CCS"" gene. Why do I get this?
Answer: As the description states, CCS stands for copper chaperone for superoxide dismutase. The ""SOD1"" appears in the record for this gene, so it comes up as a hit as well. (Look at this not as a ""bad"" hit, but realize that such a result can come in handy when you're learning about a gene: if you didn't know that there was a copper chaperone for SOD1, now you do!)